Neural Networks & Deep Learning: A.I in Biology
I'm picking up where I left off in the first half of this piece, which was published a few days ago here on @Stemsocial. Glad I got a number of valid and fantastic reactions in the comments area.
I'll proceed to discuss how the scientists behind AlphaFold and RoseTTAFold were able to bring the holy grail of biochemistry to the scientific world.
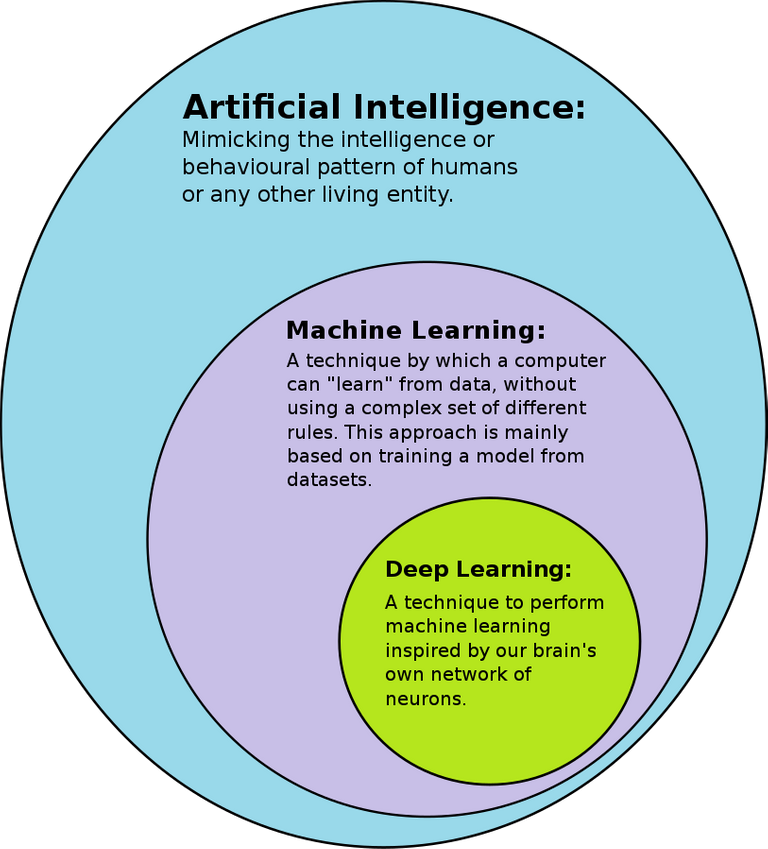
Deep learning and neural networks were used by DeepMind and the Seattle teams. The latter entails a network made up of layers of interconnected nodes (also known as "artificial neurons") that employ algorithms to replicate the way biological neurons in our brains relay signals to and from one another.
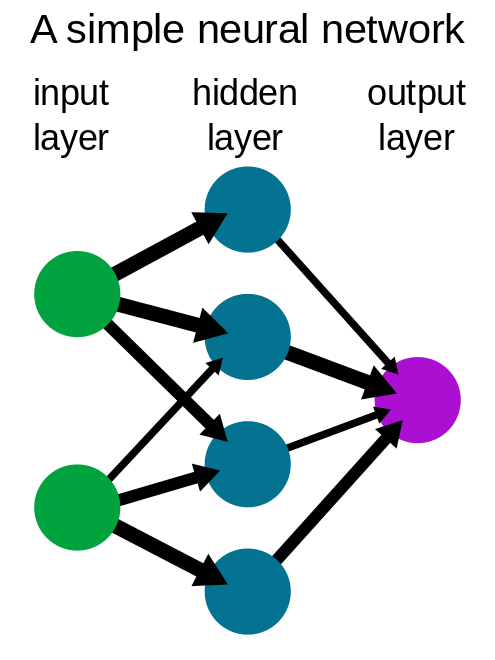
Deep learning, on the other hand, is a branch of machine learning that consists of a neural network with at least three layers of nodes. An input layer, one or more "hidden" intermediary layers, and an output layer.
One might wonder how this works. The logic here is that by combining these many layers, the program can learn to detect and identify things in a dataset. Each node in a neural network can be thought of as a miniature mathematical model that predicts an outcome using an equation. The output of the node is now the prediction, and this data will be passed as input data to another node in the neural network's next layer. The process is then repeated again and again.
To "train" the system, a researcher could utilize a set of accurately classified training data. The algorithm uses these to make predictions and check for errors against a known dataset. The data gathered from the errors can be used to fine-tune the algorithm's predictions, making it more accurate over time. When the researcher is satisfied with the algorithm, he or she might input data that the program was not trained on to see if it works properly.
This was applied to protein folding, and the training and testing data comprised correctly folded proteins. The algorithms were able to train themselves by looking at these proteins and trying to fold the proteins themselves to match what they saw. Both teams used data from the PDB (Protein Data Bank), which contains a massive library of 180k+ protein structures and amino acid sequences.
To stand out at the CASP 14 event, the teams had to do more than just put in previously-folded proteins and hope the algorithm learned how to fold more. They went the extra mile of making adjustments specific to the world of protein folding, which needed extensive biochemistry knowledge, taking use of the fact that biochemists can anticipate whether an amino acid sequence will form a coil or a flat sheet on a more general level. This knowledge was utilized to guide the neural networks' predictions.
AlphaFold employs three neural networks, each of which feeds into the other in two stages. It begins with a network that reads and folds the amino acid sequence while also adjusting the distance between pairs of amino acids in the overall structure. After that, there's a structural model network that reads everything, builds a 3D structure, and makes modifications in the end.
One of RoseTTAFolds innovation was the addition of a third neural network that tracks the location of amino acids in 3D space as the structure folds, taking into consideration both 1D and 2D data. This will be of utmost use for protein-folding experts who have begun to investigate the evolutionary history of closely related proteins. They're trying to figure out the structures of these proteins, since knowing the structure of one can help them make informed estimates about the structure of an unknown related protein.
RoseTTAFold isn't quite as exact as AlphaFold, but it works by consuming far less computing power and time than AlphaFold's, owing to the fact that RoseTTAFold's researchers didn't have access to Google's huge computational capacity to perform the calculations. RoseTTAFold can do in minutes what AlphaFold takes hours to achieve, and it can also do a host of other things that AlphaFold can't yet!
AlphaFold, for example, appears to struggle with the vital but challenging subject of protein complexes, in which numerous proteins interact and the shapes of the complexes are dependent on the specific proteins present. Understanding protein complexes is an important component of understanding proteins in general because many proteins require partners to function. RoseTTAFold can handle proteins where the amino acid chain is broken into more than one piece, and this logic can be used to investigate how different proteins interact with one another. This is extremely beneficial when it comes to analyzing complexes.
Most crucially, it is worth saying that I'm not implying these A.I. technologies will eventually replace scientists who research proteins. We wouldn't have the technology if these researchers hadn't amassed vast databases of protein structures over decades. The A.I.s are only important because they will make it easier for humans to accomplish more tasks. For example, biochemists can quickly develop and test artificial proteins, allowing them to create novel proteins that we can only fathom. Biologists will also have unprecedented access to understanding the history and nature of life! The future of biology is looking fantastic!
Thank you for taking the time to read this. I hope I was able to share what little I know with you, and I eagerly await your brilliant responses once more. Please leave them in the comments section below!
Cheers hivers!
Check out these Source Materials for detailed info
Neural network
What is deep learning and how does it work?
Accurate prediction of protein structures and interactions using a three-track neural network
Improved protein structure prediction using potentials from deep learning
Congratulations @mengene! You have completed the following achievement on the Hive blockchain and have been rewarded with new badge(s):
Your next target is to reach 42000 upvotes.
You can view your badges on your board and compare yourself to others in the Ranking
If you no longer want to receive notifications, reply to this comment with the word
STOPTo support your work, I also upvoted your post!
Check out the last post from @hivebuzz:
Support the HiveBuzz project. Vote for our proposal!
Thanks for providing this nice complement to your previous post. I knew the AlphaFold and RoseTTAFold names from elsewhere (not only from your posts) but I have never taken the time to dig deeply into what these were.
Of course machine learning will never replace scientists. machine learning is a tool, and it is important to use it wisely. Also, machine learning has its limitations. It is just that parts of our job will be different. It is more an evolution than a replacement, from my own perspective.
I sure agree with you on the perspective of the tech being more of an evolution than a replacement. They are here to ease research..
Thanks for reading through!
You are welcome!
Thanks for your contribution to the STEMsocial community. Feel free to join us on discord to get to know the rest of us!
Please consider delegating to the @stemsocial account (85% of the curation rewards are returned).
You may also include @stemsocial as a beneficiary of the rewards of this post to get a stronger support.